
Oceans are large, open habitats, where it was previously believed that the lack of obvious barriers to dispersal would result in extensive mixing, thereby preventing organisms from adapting genetically to particular habitats. It has recently become clear, however, that many marine species are subdivided into multiple populations that have evolved to thrive best under contrasting local environmental conditions. Nevertheless, we still know very little about the genomic mechanisms that enable divergent adaptations in the face of ongoing intermixing.
The project was organized into four interconnected components
NSF Outcomes Report (2023)
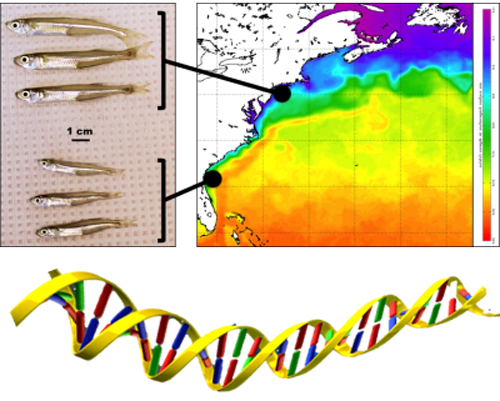
In 2017, Nina Overgaard Therkildsen and Hannes Baumann, two early career researchers from Cornell University and the University of Connecticut, formed an alliance centered on the shared fascination for a small fish. This fish, the Atlantic silverside (Menidia menidia), has been a marine vertebrate model in eco-evolutionary studies ever since experimental work in the 1970s revealed that populations from different latitudes differed genetically and showed local adaptations in many different traits along the underlying climate gradient.
But how could such a fine-scale genetic structure exist? It was long assumed that in the marine realm migration and movement with ocean currents often result in gene flow that is so predominant that most marine species should have well-mixed, genetically homogenous populations.
So Baumann, the ecologist with experience in silverside rearing, and Therkildsen, a conservation geneticist already deeply involved in deciphering the silverside genome – together hatched a plan to reveal the genomic underpinnings of local adaptation in this famous model system. They proposed a combination of field collections and rearing experiments organized in four hierarchical parts they called SCAN, MAP, EVALUATE, and TEST.
More than five years later, we look back on a number of discoveries and groundbreaking experiments. And yet, it still feels like we have only scratched the surface of how evolution and genomes shape nature around us. The project demonstrated the existence of extreme levels of structural genetic diversity and hundreds of linked genes in M. menidia (Wilder et al. 2020, Tigano et al. 2021). Central to the main question was the discovery of massive chromosomal inversions in the silverside genome, which likely facilitate local adaptation in this species (Akopyan et al. 2022). We also worked on and published a comprehensive, non-redundant reference transcriptome for the Atlantic silverside (Therkildsen & Baumann 2020) that has since become an important resource for follow-up studies (Jacobs et al. in review).
In one of the most memorable and daring experimental efforts, our two teams drove for 36 hours to sample spawning-ripe silversides at Jekyll Island in Georgia, transport them live to Connecticut and then collect ripe spawners from another site at the south shore of Long Island (Patchogue). The reciprocal, individual and population F1 crosses were reared to maturity and then spawned again to obtain and rear F2 offspring, which then allowed quantitative trait mapping and evaluation of the role and ubiquity of inversions.
Overall, this project produced a treasure trove of data that will continue to yield insights into the genomic architecture enabling local adaptation in silversides for several years to come. However, along with a groundswell of other work, this project has already and squarely rendered the notion outdated that marine populations are genetically homogeneous. They often are clearly not, and chromosomal inversions are likely a major reason why such genetic population structure and local adaptations persist in the wild.
Both PI’s and students have given over 20 presentations, seminars, and invited lectures, and the project has so far resulted in 7 high-profile publications, with another manuscript still in review. The project has supported training and career development for 8 graduate students, 3 post-doctoral female researchers, and 1 technician.
It broadly succeeded in establishing the Atlantic silverside as a model system for advanced ecological, evolutionary, and molecular approaches, while promoting the wonder and amazement about evolutionary principles in marine organisms.
Selected references
Akopyan M, Tigano A, Jacobs A, Wilder AP, Baumann H, Therkildsen NO (2022) Comparative linkage mapping uncovers recombination suppression across massive chromosomal inversions associated with local adaptation in Atlantic silversides. Molecular Ecology 31:3323-3341
Jacobs, A., Velotta, J.P., Tigano, A., Wilder, A.P., Baumann, H., and Therkildsen, N.O. (2023) Temperature-dependent gene regulatory divergence underlies local adaptation with gene flow in the Atlantic silverside. Evolution (in review)
Therkildsen NO, Baumann H (2020) A comprehensive non-redundant reference transcriptome for the Atlantic silverside Menidia menidia. Marine Genomics 53:100738
Tigano A, Jacobs A, Wilder AP, Nand A, Zhan Y, Dekker J, Therkildsen NO (2021) Chromosome-level assembly of the Atlantic silverside genome reveals extreme levels of sequence diversity and structural genetic variation. Genome Biology and Evolution 13:evab098
Wilder AP, Palumbi SR, Conover DO, Therkildsen NO (2020) Footprints of local adaptation span hundreds of linked genes in the Atlantic silverside genome. Evolution Letters 4:430-443






