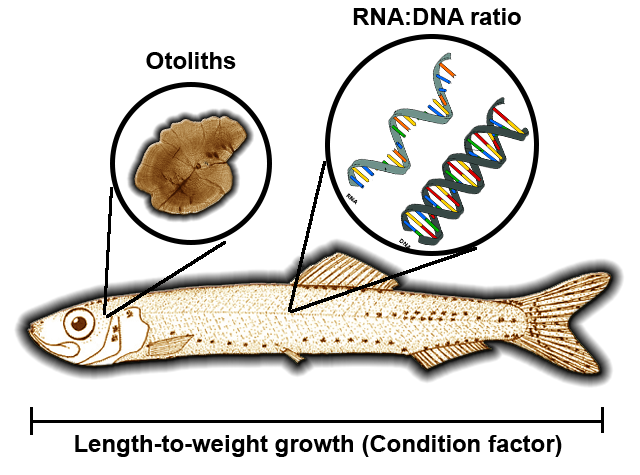
Peck, M. A., H. Baumann, C. Clemmesen, J. P. Herrmann, M. Moyano, and A. Temming 2015. Calibrating and comparing somatic-, nucleic acid-, and otolith-based indicators of growth and condition in young juvenile European sprat (Sprattus sprattus). Journal of Experimental Marine Biology and Ecology 471:217-228.
Month: June 2015
[Presentation] H. Baumann talks at the 3rd Ocean Acidification PI Meeting in Woods Hole, MA
“Plastic and evolutionary responses to ocean acidification: navigating the difficult terrain between unfounded pessimism, optimism, and impossible tasks”
Woods Hole Oceanographic Institution, 11 June 2015
The talk is publicly accessible on Prezi
[Lab News] Seining in Mumford Cove
Here are some pictures from one of our first beach seining trips to Mumford Cove, CT (16 May 2015)
[Lab News] Our new rearing system is operational!
– The maiden voyage –
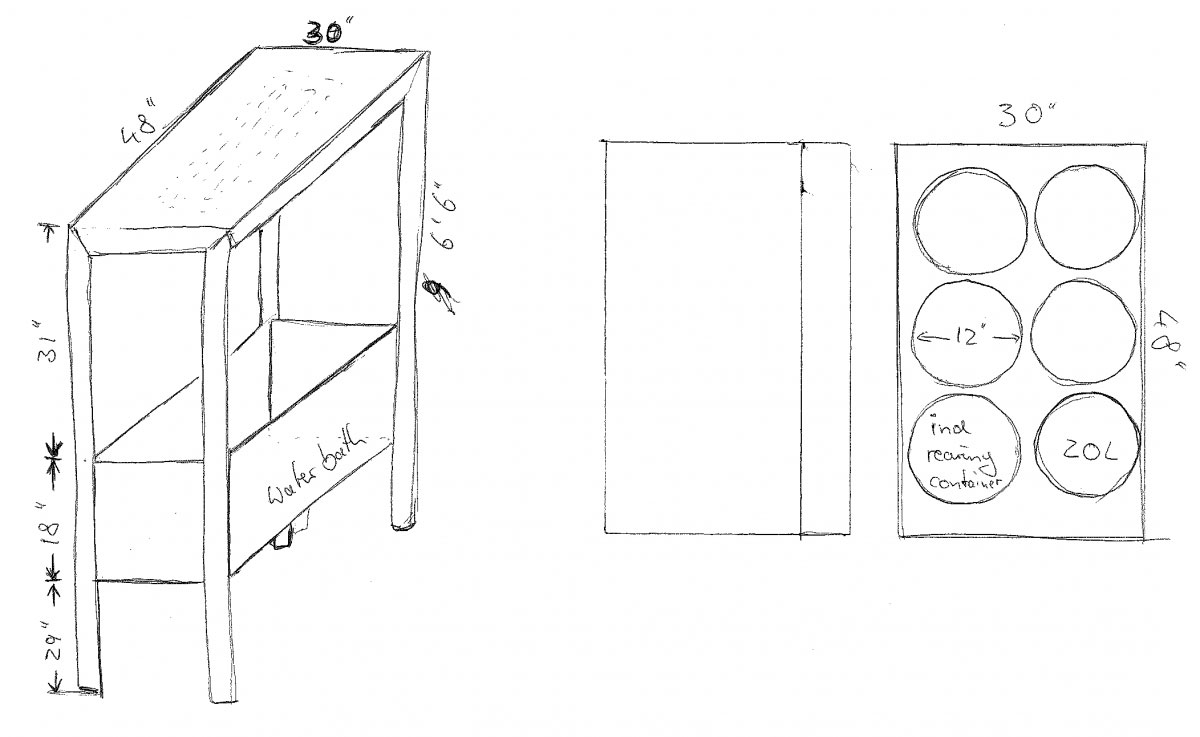
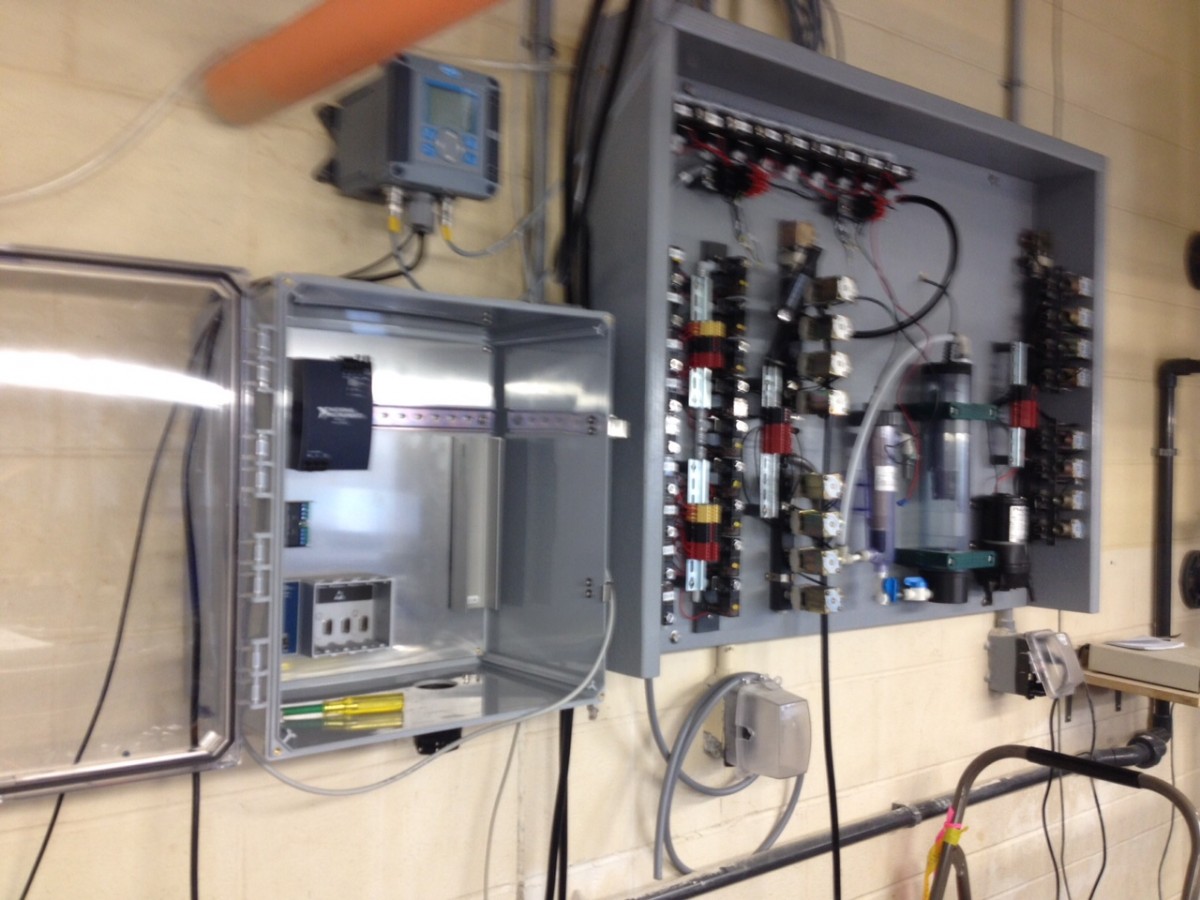
Although it still lacks a proper name, our experimental system to rear larval fish under different temperature, CO2, and oxygen conditions has finally started it’s first real trial with newly fertilized silverside embryos. The system consists of 9 independent units to allow any factorial 3 x 3 combination of rearing conditions. Each unit has a sealed main tank (400L) in which up to six individual rearing containers (20L) can be placed. Water samples from each unit are sequentially pumped past two wall-mounted oxygen and pH sensors, which feed their data into a computer program called LabView (NI), which in turn triggers solenoid valves to add CO2, nitrogen, or air to each system. Although straightforward in principle, the practice of putting all of this together is definitely more complicated and the devil in the detail. All told, the construction took us ~ 8 months, but we hope to be using this system for years from now.
Thumbs up to the many, many persons that were instrumental to the success of this; Paul Grecay and Timothy Targett (University of Delaware) for giving us the crucial inspiration about the general design of a system like that. Gary Grenier and Bob Dziomba from the machine shop for building the big pieces and thinking ahead of details that we certainly would have missed. Charlie Woods for his excellent help and assistance in the Rankin Lab, from plumbing to electrical to simply cheering us up. Dennis Arbige for taking on the tedious wiring of the solenoids without blinking an eye. Finally, many thanks to John Hamilton who’s excellent knowledge of LabView and great teaching skills helped Chris to become a LabView wiz in a matter of weeks!
Ready. Set. Go!