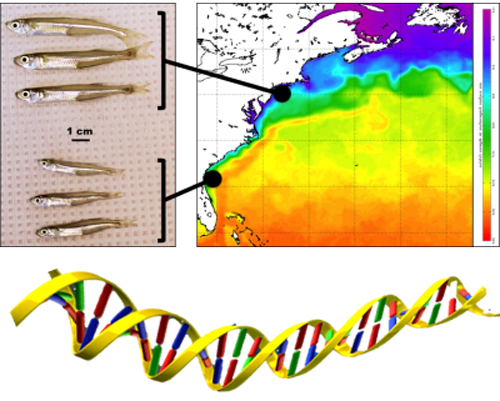
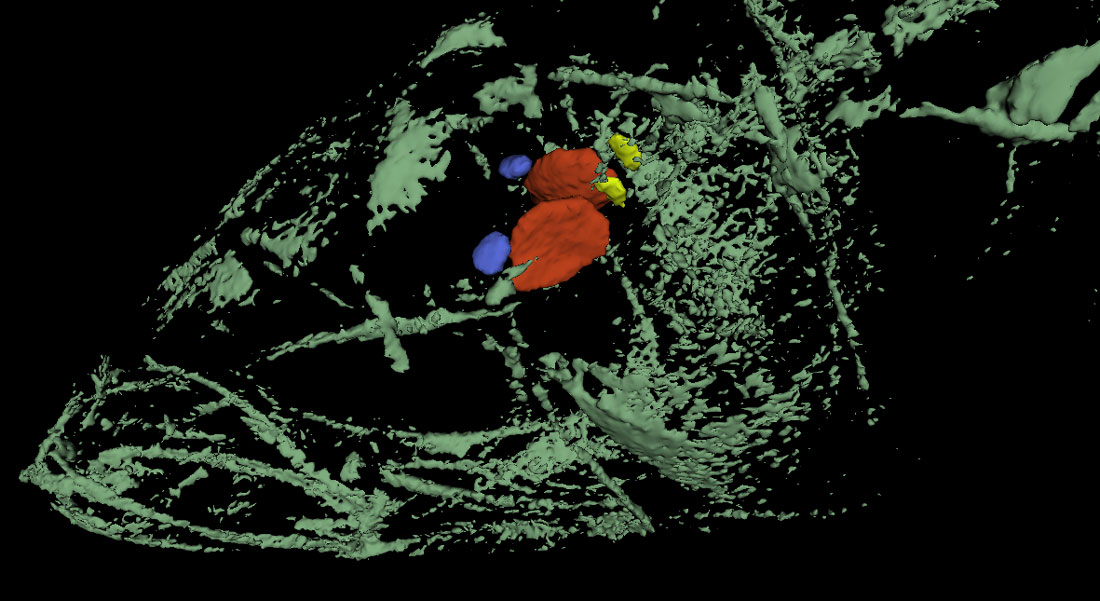
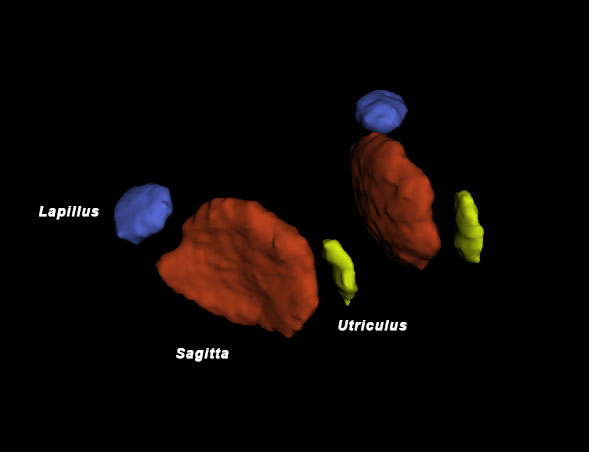
26 March 2018. Today we got our first glimpse of an incredible new way of imaging the inner calcified structures of a fish body, particularly the ear bones (otoliths), of which every teleost fish has six, three on each side inside the skull. Otoliths are long known to fish ecologists for their properties to record and store information about a fish’s age, growth and habitat. With an adult Atlantic silverside, Hannes visited John Shepherd, facilities scientist and member of the Goldhamer lab at UConn, Storrs (Biology Physics Building), who showed us the use of a new, state-of-the-art micro CT-scanner (IVIS). Turns out, the system effortlessly imaged all six otoliths inside of the fish’s head. Later in the year, we will use the technique to image silversides reared at contrasting CO2 conditions to see whether they differ in their otolith size, volume, and structure. Thank you, John, for this truly inspiring demonstration!
Month: March 2018
[New publication] Brachiopods resilient to global change
Congratulations to Emma Cross to her new publication in Global Change Biology today!

Read the full press release by the British Antarctic Survey
- Cross et al. 2018. A 120-year record of resilience to environmental change in brachiopods.
Global Change Biology published online 14 March 2018
[Research news] New NSF grant to study silverside genes!
We are overjoyed to announce that NSF is funding a new and collaborative research project to look at the genomic underpinning of local adaptation in the Atlantic silverside! Check out below for a first glimpse of the project website.

Oceans are large, open habitats, where it was previously believed that the lack of obvious barriers to dispersal would result in extensive mixing, thereby preventing organisms from adapting genetically to particular habitats. It has recently become clear, however, that many marine species are subdivided into multiple populations that have evolved to thrive best under contrasting local environmental conditions. Nevertheless, we still know very little about the genomic mechanisms that enable divergent adaptations in the face of ongoing intermixing.