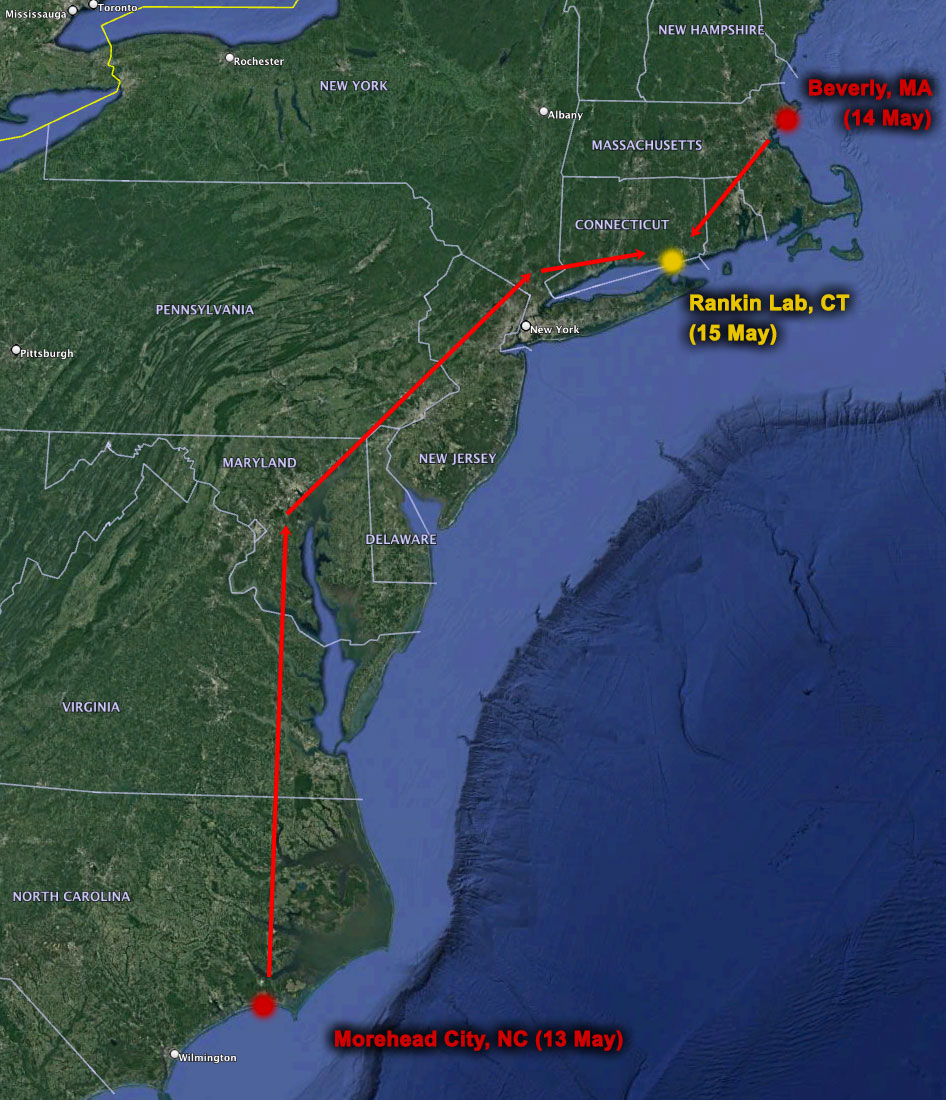
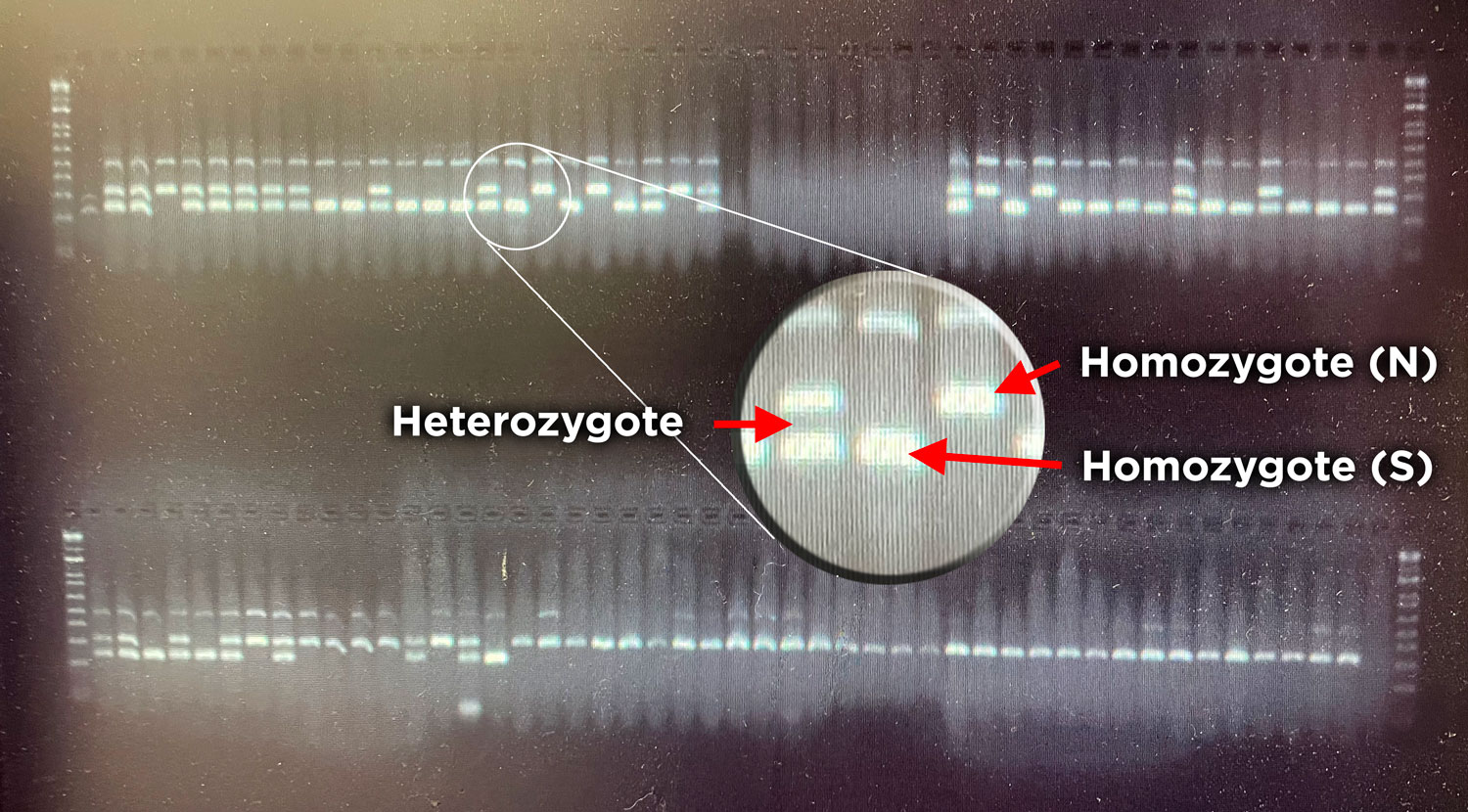
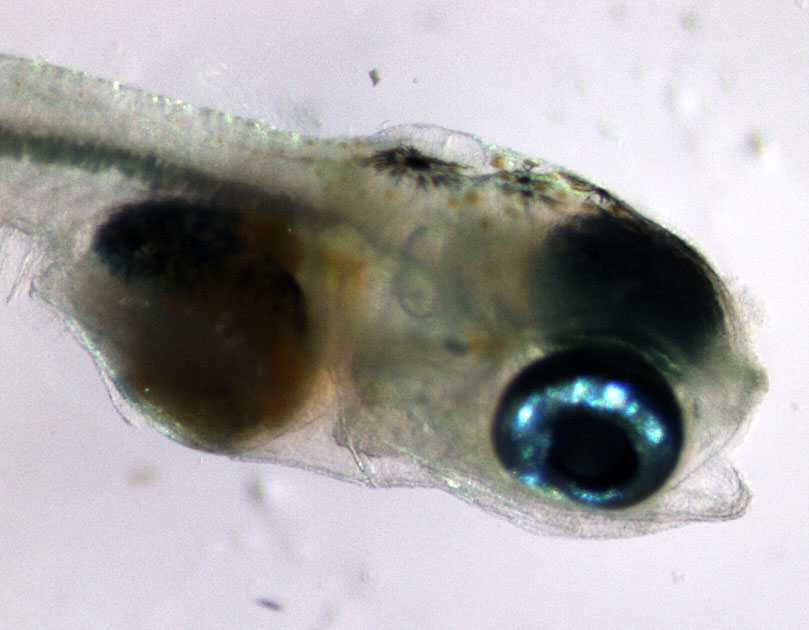
15 May 2022. A full, blood red moon rises over Pine Island this Sunday evening. The sight makes not just humans swoon – its pull extents underwater to all kinds of critters that take it as cue for reproduction. Critters just like the Atlantic Silverside, which once again we pursue this season to extract more of its genomic 'secrets'.
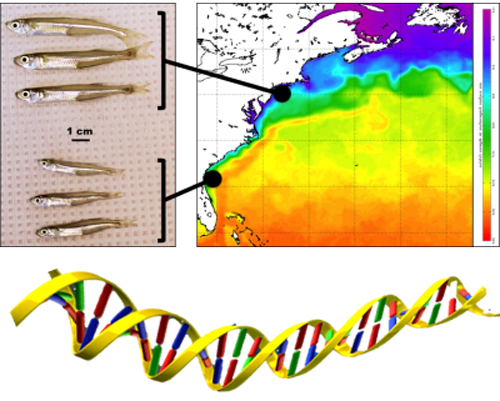
Specifically, it is this weekend that we embark on yet another ambitious road trip to find and sample spawning-ripe silversides from two very far apart places: Morehead City, North Carolina and Beverly, Massachusetts. The goal: transport spawners live from each population to UConn's Rankin Seawater lab and produce calculated crosses that will allow studying the role of genomic inversions in local adaptation.
The crew this time are Maria Akopyan and Jessica Rick from Cornell University, along with Lucas Jones and Hannes Baumann from UConn. Big shout-out to Tara Duffy for her help with beach seining at Beverly, MA. During the spawning event on May 15th, Nina Therkildsen also joined the efforts. The design and experiment are part of Jessi's successful NSF post-doctoral fellowship proposal, which the whole UConn-Cornell silverside team supports.
Click through the pictures below to retrace the steps of an exhausting but so far successful effort. Fingers crossed that all goes well during the next weeks, when the fish need to hatch, survive and grow, so they can be assessed for their traits.