
Well done, Dr. Murray! We are all so very proud of you!

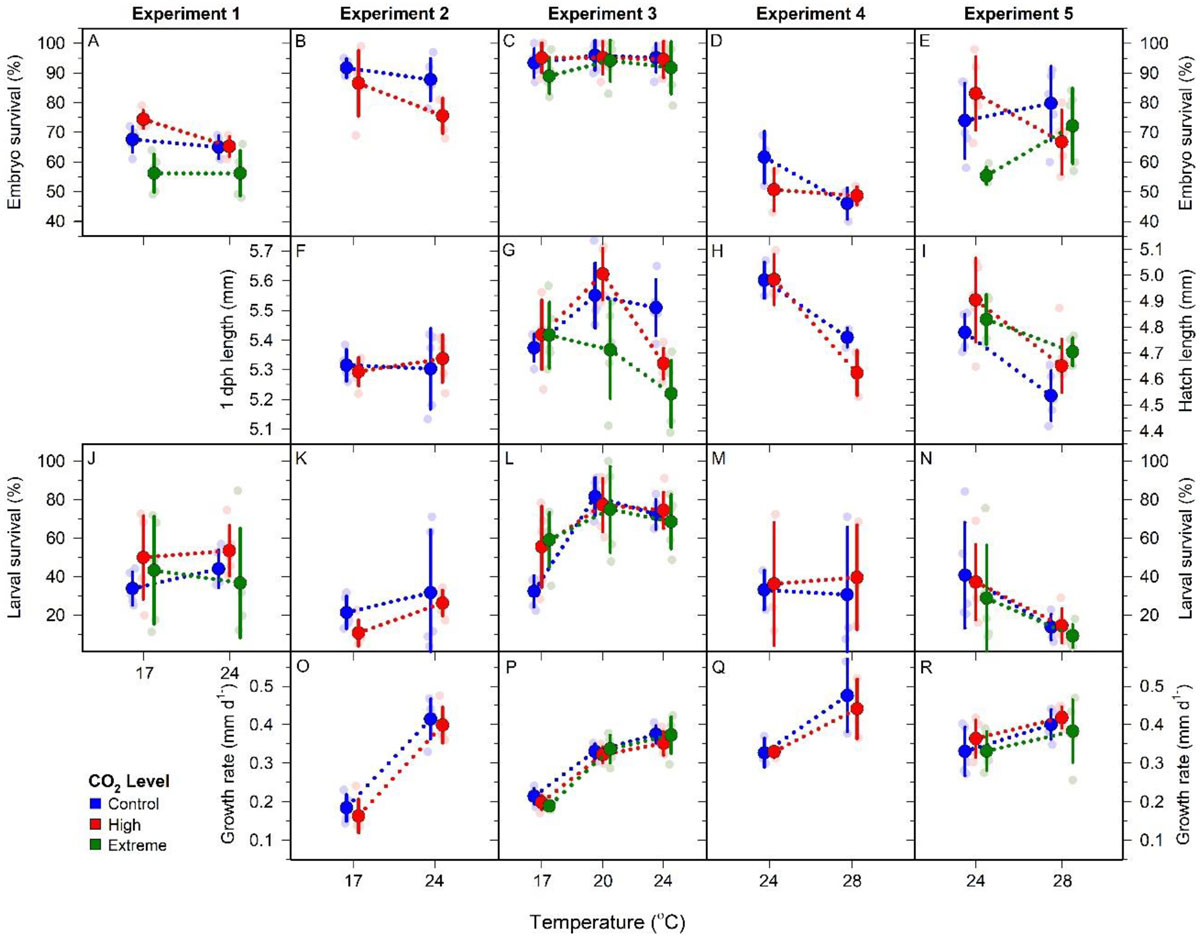
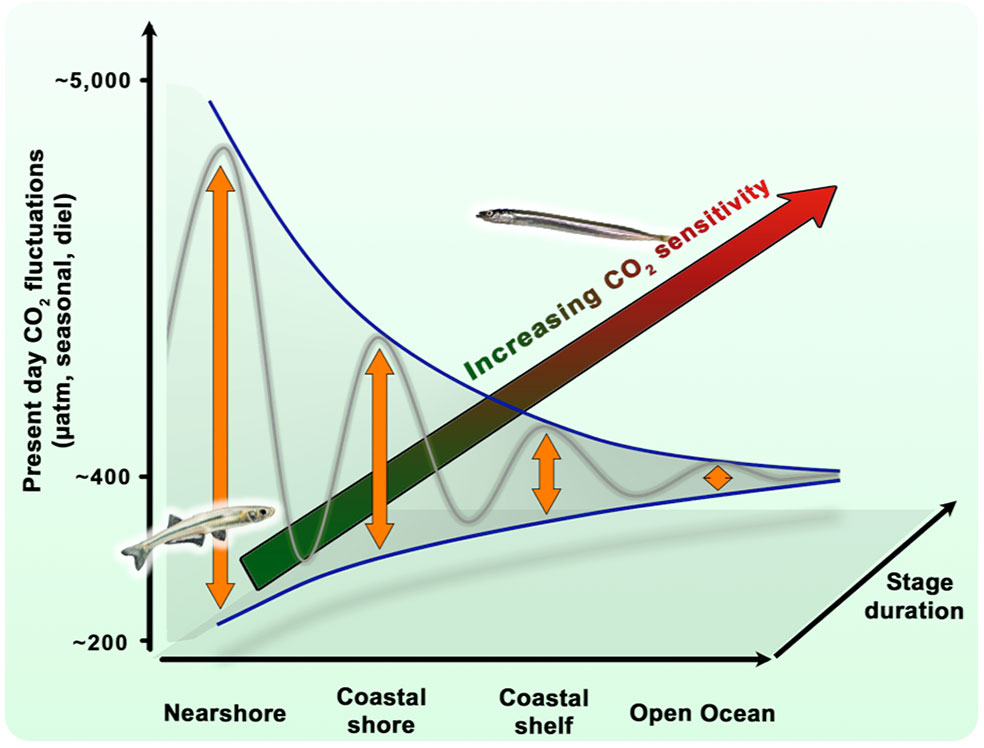
Baumann, H., Cross, E.L., and Murray, C.S. Robust quantification of fish early life CO2 sensitivities via serial experimentation. Biology Letters 14:20180408

The goal of the sandlance cruise was to collect running ripe males and females to do a fertilization via strip spawning. Emma and I were a bit doubtful at first because we got less than 10 sandlance on the first two trolls. However, things got much better by the afternoon, and our most successful trawl caught 147 sand lance. I helped out with the fertilization and deploying the trawl, two things I have never done before. The most exciting part of the day was getting to see humpback whales. Usually they are in the distance but today they were right next to the boat. Everyone on board said that this never happens and it was very unusual so I felt very lucky to have seen whales at such a close proximity.”
Overall, the trip was a huge success and it was very refreshing to see everything go as planned. The only downside to the day was driving back home through a snowstorm. I later found out that there was a 73% fertilization success and we got 27,000 embryos for Emma’s experiment. I am very grateful to have gotten the opportunity to help out on this sampling cruise and am looking forward to doing this again in the future!


12 October 2018. This year, the Department of Marine Sciences at UConn Avery Point has conducted his first graduate course on physical and biological oceanographic methods, which culminated in a two day research cruise aboard the newly stretched R/V Connecticut. The cruise sampled stations from Eastern Long Island Sound all the way out the continental shelf, deploying CTD’s, sediment corers and grabs, as well as zooplankton and nekton nets. Callie and Hannes from the Baumann lab were part of the fun!
Check out some of the action in the youtube clip below.

This technique involves placing live fish in a fluorescent mitrochondrial stain for 5 minutes before imaging different areas of the fish under a dissecting microscope equipped with an epiflourescence light source. This allowed us to visualize small sense organs called neuromasts located in tubular canals in the head, trunk and tail, which form the fish sensory lateral line system used to detect water flows.
Congrats, Chris, to the second chapter published!


The particularly intimate format of the Gordon Research Conference was wonderfully conducive to listening to groundbreaking science in form of keynote lectures and posters and to network with colleagues from all over the world. While Hannes gave a keynote lecture about experimental progress in assessing fish sensitivity to marine climate change, Chris, Emma, Jimmy and Hans presented their research all throughout the week during the poster sessions. The beautiful setting of the conference in New Hampshire’s White Mountains and the relaxed atmosphere were all contributing to one of the most unique conference experiences all year.
Talks and posters presented:



Ever since attending the American Fisheries Society conference in 2014, I’ve wanted to go to another fish-focused conference. I was lucky enough to attend the 42nd annual Larval Fish Conference this year in Victoria, British Columbia, and it surpassed all my expectations. The week started off with a larval fish identification workshop where we got to learn techniques from renowned larval fish experts (and see some really cool fish larvae!). The talks were impressive and thought-provoking, providing many new ideas for research and how to give an engaging talk. My favorite part was meeting all the larval fish ecologists whose publications I’ve been reading for my thesis. I spent most of my evenings exploring Victoria with other grad students attending the conference and left with many new friends from institutes all over the world! The trip ended with a whale watch, where we saw a pod of 5 Orcas. Overall, the Larval Fish Conference was a great experience that I hope to someday attend again!
Oral presentations: