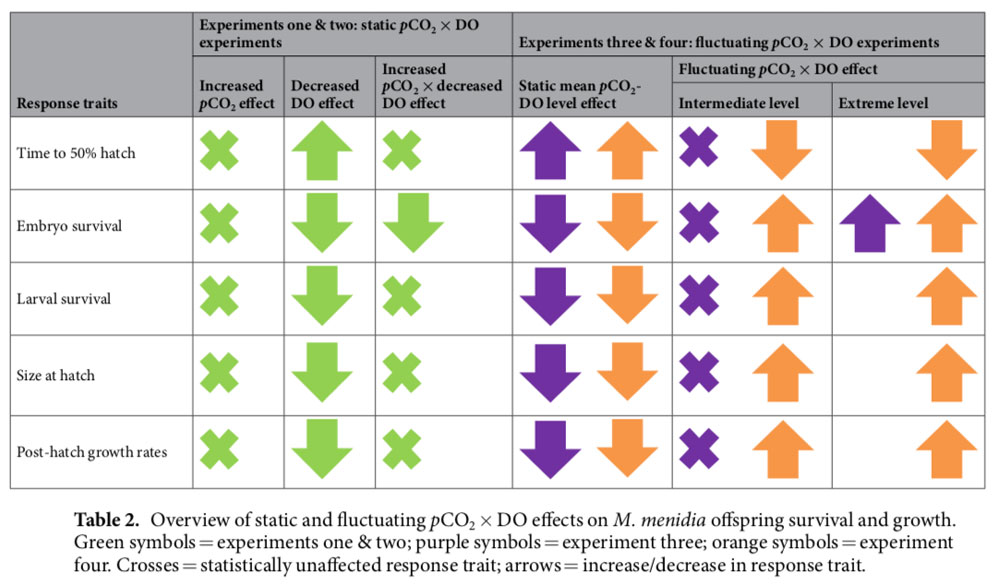
27 July 2020. Big and proud congratulations to Chris Murray, who published his last big chunk of data from his PhD research on the effects of marine climate change on coastal marine fish. The publication in PLOS One synthesized 3 years of multiple, long-term experiments on Atlantic silversides (Menidia menidia) demonstrating consistent negative growth effects on high CO2 conditions. However, sometimes it takes more than just looking at means and standard deviations to elucidate these effects. Hence, in this paper, shift functions analyzing the different percentiles of distributions are employed.
Murray, C.S. and Baumann, H. (2020) Are long-term growth responses to elevated pCO2 sex-specific in fish? PLOS One 15:e0235817
The publication was featured in UConn Today “UConn Research: More Carbon in the Ocean Can Lead to Smaller Fish”
By Elaina Hancock
As humans continue to send large quantities of carbon into the atmosphere, much of that carbon is absorbed by the ocean, and UConn researchers have found high CO2 concentrations in water can make fish grow smaller.
Researchers Christopher Murray PhD ’19, now at the University of Washington, and UConn Associate Professor of Marine Sciences Hannes Baumann have published their findings in the Public Library of Science (PLoS One).
“The ocean takes up quite a bit of CO2. Estimates are that it takes up about one-third to one-half of all CO2 emissions to date,” says Murray. “It does a fantastic job of buffering the atmosphere but the consequence is ocean acidification.”
Life relies on chemical reactions and even a slight change in pH can impede the normal physiological functions of some marine organisms; therefore, the ocean’s buffering effect may be good for land-dwellers, but not so good for ocean inhabitants.
Baumann explains that in the study of ocean acidification (or OA), researchers have tended to assume fish are too mobile and tolerant of heightened CO2 levels to be adversely impacted.
“Fish are really active, robust animals with fantastic acid/base regulatory capacity,” says Murray. “So when OA was emerging as a major ocean stressor, the assumption was that fish are going to be OK, [since] they are not like bivalves or sea urchins or some of the other animals showing early sensitivities.”
The research needed for drawing such conclusions requires long-term studies that measure potential differences between test conditions. With fish, this is no easy task, says Baumann, largely due to logistical difficulties in rearing fish in laboratory settings.
“For instance, many previous experiments may not have seen the adverse effects on fish growth, because they incidentally have given fish larvae too much food. This is often done to keep these fragile little larvae alive, but the problem is that fish may eat their way out of trouble — they overcompensate – so you come away from your experiment thinking that fish growth is no different under future ocean conditions,” says Baumann.
In other words, if fish are consuming more calories because their bodies are working harder to cope with stressors like high CO2 levels, a large food ration would mask any growth deficits.
Additionally, previous studies that concluded fish are not impacted by high CO2 levels involved long-lived species of commercial interest. Baumann and Murray overcame this hurdle by using a small, shorter-lived fish called the Atlantic silverside so they could study the fish across its life cycle. They conducted several independent experiments over the course of three years. The fish were reared under controlled conditions from the moment the eggs were fertilized until they were about 4 months old to see if there were cumulative effects of living in higher CO2 conditions.
Murray explains, “We tested two CO2 levels, present-day levels and the maximum level of CO2 we would see in the ocean in 300 years under a worst-case emissions scenario. The caveat to that is that silversides spawn and develop as larvae and early juveniles in coastal systems that are prone to biochemical swings in CO2 and therefore the fish are well-adapted to these swings.”

The maximum CO2 level applied in the experiments is one aspect that makes this research novel, says Murray,
“That is another important difference between our study and other studies that focus on long-term effects; almost all studies to date have used a lower CO2 level that corresponds with predictions for the global ocean at the end of this century, while we applied this maximum level. So it is not surprising that other studies that used longer-lived animals during relatively short durations have not really found any effects. We used levels that are relevant for the environment where our experimental species actually occurs.”
Baumann and Murray hypothesized that there would be small, yet cumulative, effects to measure. They also expected fish living in sub-ideal temperatures would experience more stress related to the high CO2 concentrations and that female fish would experience the greatest growth deficits.
The researchers also used the opportunity to study if there were sex-determination impacts on the population in the varying CO2 conditions. Sex-determination in Atlantic silversides depends on temperature, but the influence of seawater pH is unknown. In some freshwater fish, low pH conditions produce more males in the population. However, they did not find any evidence of the high CO2 levels impacting sex differentiation in the population. And the growth males and females appeared to be equally affected by high CO2.
“What we found is a pretty consistent response in that if you rear these fish under ideal conditions and feed them pretty controlled amounts of food, not over-feeding them, high CO2 conditions do reduce their growth in measurable amounts,” says Murray.
They found a growth deficit of between five and ten percent, which Murray says amounts to only a few millimeters overall, but the results are consistent. The fish living at less ideal temperatures and more CO2 experienced greater reductions in growth.
Murray concludes that by addressing potential shortcomings of previous studies, the data are clear: “Previous studies have probably underestimated the effects on fish growth. What our paper is demonstrating is that indeed if you expose these fish to high CO2 for a significant part of their life cycle, there is a measurable reduction in their growth. This is the most important finding of the paper.”
This work was funded by the National Science Foundation grant number OCE #1536165. You can follow the researchers on Twitter @baumannlab1 and @CMurray187.